- Published on
Deploying AI in Agriculture: How we built a Semi-Supervised ViT pipeline for ICAR
Agriculture is at the heart of many economies, and early detection of crop diseases can save entire harvests and livelihoods. Leveraging advances in AI, I developed a semi-supervised learning pipeline combining EfficientNet and Vision Transformers to accurately classify multiple crop diseases from field-collected images.
This project, supported by a joint initiative between the Indian Council of Agricultural Research (ICAR) and the National Institute of Technology Karnataka (NITK), achieved a 95.7% classification accuracy despite working with a limited dataset (~500 samples) of real-world images of pomegranate leaves and fruit. The final model was optimized for edge deployment using ONNX quantization, enabling practical use in field diagnostics.
Disclaimer: This project was carried out under a funded collaboration mediated by NITK with ICAR. While ICAR provided support for the initiative, all project development and implementation were conducted by our team at NITK, without direct engagement with ICAR personnel.
- The Goal: Integrating AI for Real-World Pomegranate Crop Monitoring
- Challenges and limitations
- An End-to-End Vision Pipeline implementation
- Conclusion
The Goal: Integrating AI for Real-World Pomegranate Crop Monitoring
Pomegranate cultivation plays a vital role in the agricultural economy of several Indian states, notably Maharashtra, Karnataka, Andhra Pradesh, and Gujarat. Accordingf to ICAR, pomegranate crops in these areas are frequently threatened by a range of diseases (primarily bacterial and fungal) that can severely reduce yield and fruit quality if not detected early.
Our goal was to build a deep learning system capable of accurately identifying multiple pomegranate diseases from images captured in real farming environments, using a highly limited dataset of approximately 500 field-collected images of leaves and fruits. These images were taken under varying lighting conditions, angles, and backgrounds using smartphones on the field.

The dataset we worked with includes images representing fungal diseases such as Alternaria and Anthracnose, bacterial infections like Bacterial blight, as well as healthy leaf and fruit samples. These categories capture the diversity and complexity of field conditions faced by farmers.
Challenges and limitations
Highly Limited dataset
Our dataset consisted of only ~500 images each of leaves and fruit. With such a small volume of labeled data, training deep models without overfitting was a significant challenges
Irregularity in size and features
Images varied wildly in resolution, aspect ratio, and focus. Since they were captured in uncontrolled environments, many had motion blur, shadows, or inconsistent framing—making feature extraction harder and reducing overall model reliability without preprocessing and augmentation.
⚠️ Background Bias in Class Labels
A major hidden challenge we uncovered mid-way was background bias.
- Healthy pomegranate samples were almost exclusively captured on the tree, surrounded by leafy green backgrounds.
- Bacterial infection images came from a uniform source with consistent backgrounds.
- Fungal disease images had more diverse and complex surroundings.
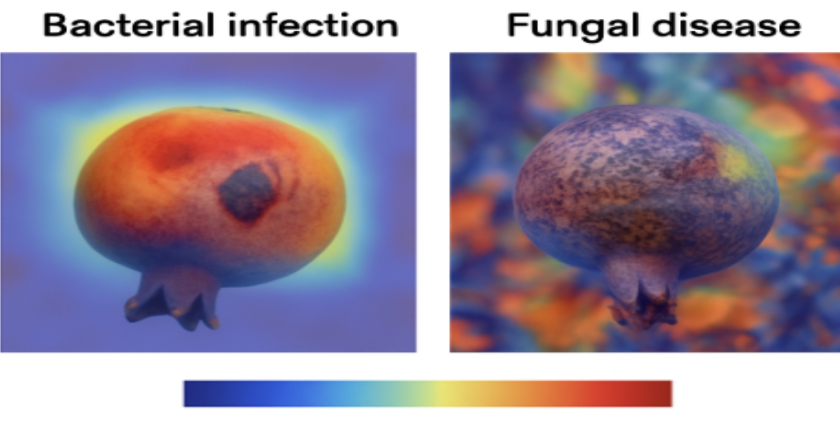
We noticed unexpectedly high validation accuracy in early CNN models and suspected overfitting. Using Grad-CAM, we discovered the models were often focusing on background textures rather than the fruit or leaves themselves.
An End-to-End Vision Pipeline implementation
1. Preprocessing: Augmentations and Background Removal
Data Augmentations: Rotations and randomized resizing were used to simulate field variability and successfully expand the size of the dataset.
Background removal: To combat background-based shortcuts, we applied a lightweight segmentation model to isolate the fruit or leaf. The resulting mask was overlaid on the image to de-emphasize or whiten out distracting background textures.

2. Model Architecture
To balance feature richness and model efficiency, we used a hybrid architecture:
EfficientNet-B0 served as the CNN backbone for spatial feature extraction.
Features were passed to a Vision Transformer (ViT) encoder, which applied attention mechanisms to identify relevant regions—particularly useful for spotting localized infections.
A final softmax classifier used to classify the fruits or leaves into one of the three classes - Bacterial, Fungal, Healthy
This combination allowed us to capture both local patterns (via CNN) and global dependencies (via ViT), leading to robust classification across disease types.
3. Semi Supervised Learning
With only ~500 labeled images, we turned to semi-supervised learning (SSL) to make use of unlabeled field data and implemented a FixMatch-inspired framework:
- For unlabeled images, the model generated pseudo-labels using weak augmentations.
- The same images were strongly augmented and passed back to the model.
- A consistency loss ensured the model produced similar predictions across both versions.
We combined this with a supervised cross-entropy loss on the labeled data. The final loss was:
This approach significantly improved generalization without needing more expert-labeled data.
4. Export to ONNX
To prepare the model for deployment on low-resource devices, we exported the final trained model to ONNX format by:
- Tracing the PyTorch model with a dummy input using torch.onnx.export.
model.eval()
torch.onnx.export(model, dummy_input, "model.onnx", input_names=["input"], output_names=["output"])
- Applying post-training quantization to reduce model size and inference latency—ideal for mobile and edge use.
quantize_dynamic("model.onnx", path, weight_type=QuantType.QInt8)
This quantized ONNX model is now ready for integration into ICAR’s diagnostic tool for real-world field deployment. An ONNX runtime may then be used irrespective of framework to test as:
session = onnxruntime.InferenceSession(quantized_model_path)
inputs = {session.get_inputs()[0].name: dummy_input.numpy()}
outputs = session.run(None, inputs)
Conclusion
The potential of deep learning in real world use cases remains relatively untapped. Even with limited, noisy, and non-standard data, high accuracy may be obtained by combining semi-supervised learning, careful preprocessing, and a hybrid EfficientNet–ViT architecture. Equally important is ensuring that these models are usable beyond research environments into a lightweight and framework-agnostic solutions that can be deployed across a wide range of devices. This reinforces a key message:
High-performance AI edge systems don’t always require massive datasets—just smarter engineered pipelines.
Enjoyed this post? Subscribe to the Newsletter for more deep dives into ML infrastructure, interpretibility, and applied AI engineering or check out other posts at Deeper Thoughts